How to run the notebooks
Option 1. Using BiocWorkshop (Preferred)
If you are attending the workshop at Bioconductor:
- Open BiocWorkshops in a new tab.
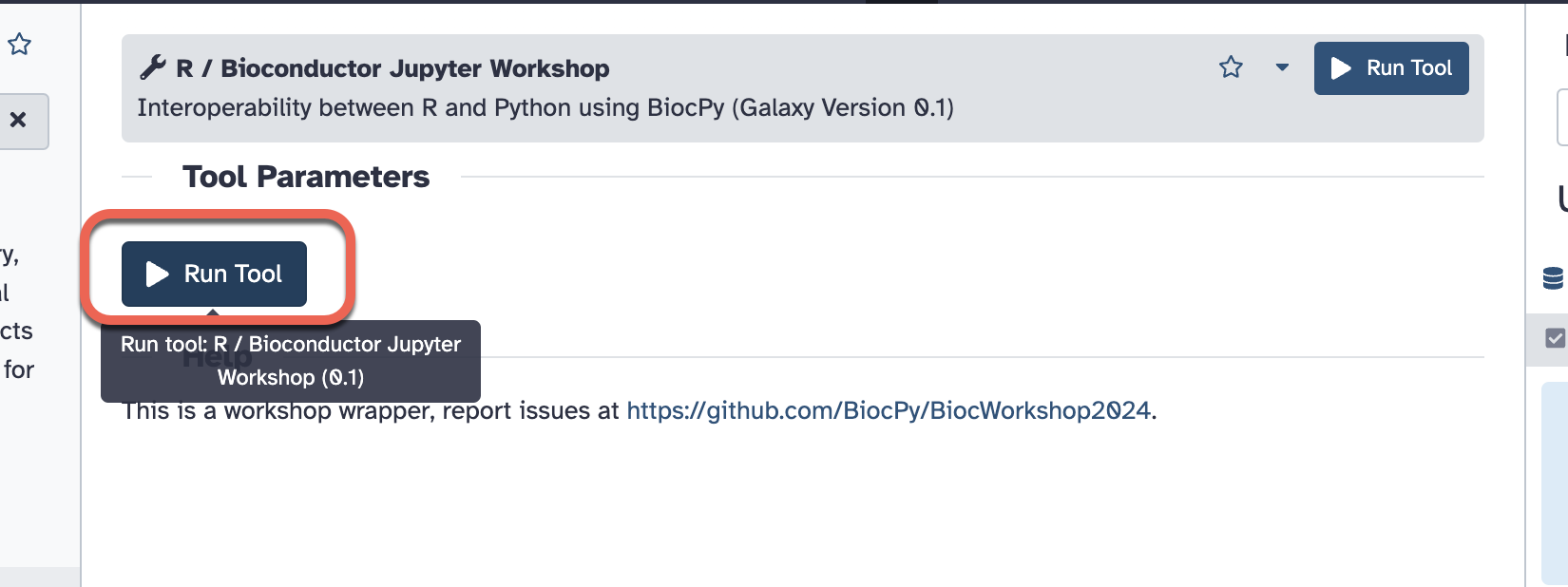
- Select “R/Bioconductor Jupyter Workshop” as shown in the screenshot.

- Click the “Run the tool”.
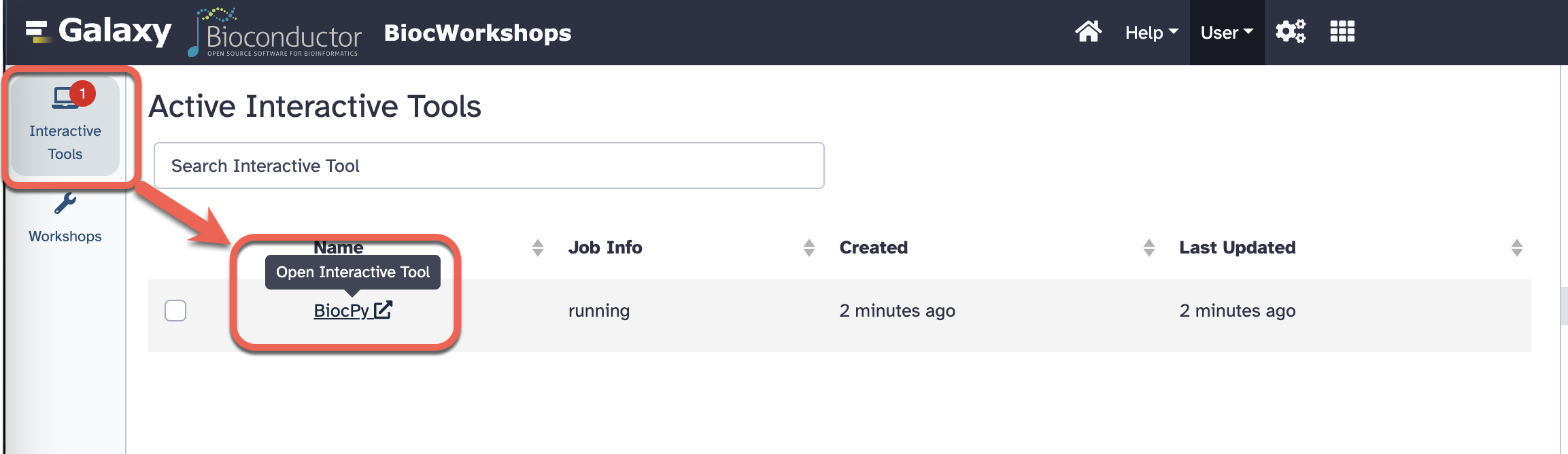
- Click on “Interactive Tools” in the left sidebar to check on the status of the session. Once the session is ready, click on the “BiocPy” to open the Jupyter notebook.
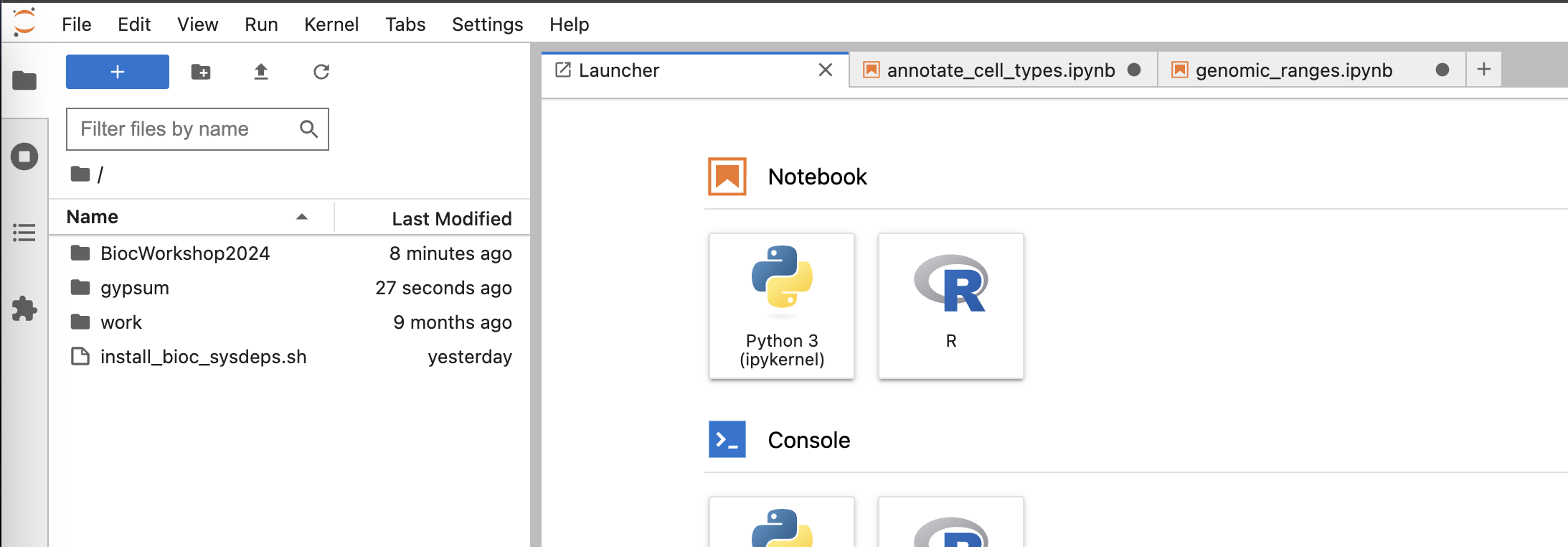
- You should be able to see the BiocWorkshop repository cloned into the session.
Option 2: Using Google Colab
To open the session in Google Colab:
- Open Google colab in a new tab.
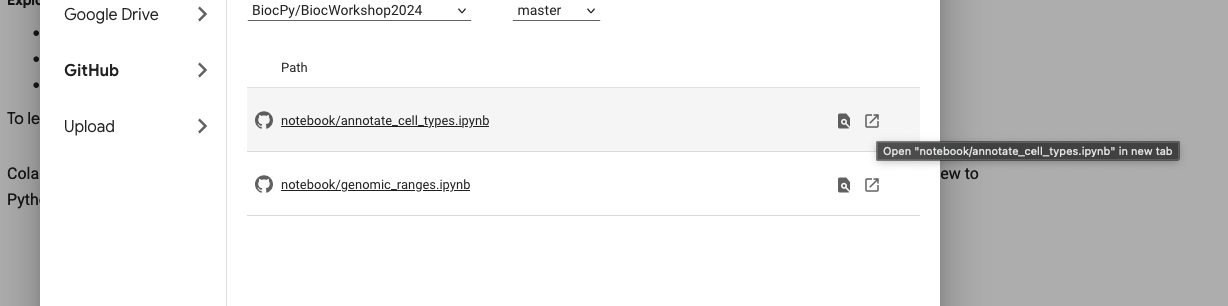
- On the page, it should open up a dialog to “Open notebook” as shown in the screenshot below.
- If no dialog appears, go to File -> Open Notebook from the menu (in the top left).
- Choose Github and enter the repository url for the workshop:
https://github.com/BiocPy/BiocWorkshop2024. This will automatically find the python notebooks available in the repository.
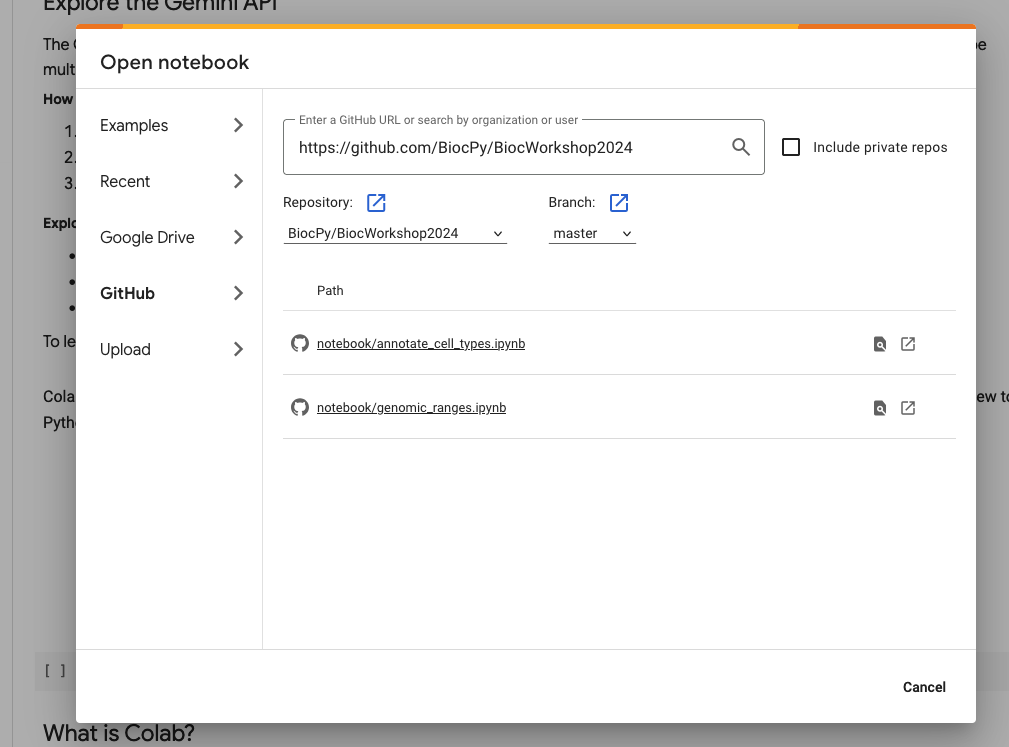
- Choose the open icon to explore this notebook
Important
This process does not download the RDS file available in the repository. Folks might have to manually download this to their Google Colab sessions.
Option 3: Run notebooks locally
If you want to run locally, please clone the repository and install the python packages used for the workshop.
git clone https://github.com/BiocPy/BiocWorkshop2024
cd BiocWorkshop2024
# Assuming python is available
# You are free to use mamba, conda or virtualenv's
pip install -r requirements.txt
# Start the jupyter server
jupyter labThen checkout the notebook directory that contain Jupyter notebooks.
